
Protein Labeling
Over the last decades, fluorescence microscopy has become an indispensable tool in many scientific fields such as cell biology and medicine. It provides a greater contrast and sensitivity compared to conventional transmitted-light microscopy. Use of spectrally distinct fluorescent markers enables simultaneous visualization of different targets within the same sample. Superresolution microscopy (e.g. STED, dSTORM) and FRET are some of the most popular fluorescence techniques providing unique insights into cellular processes, distribution and structural dynamics of proteins on the lower nanometer scale, far below the natural diffraction limit of light.
There are several approaches how to fluorescently label a protein of interest (POI), namely direct conjugation with a fluorescent organic dyes, fusion of POI with self-labeling tags (SLP) or fluorescent proteins, and immunolabeling. For all of them, it is crucial to prevent any alternation of the protein’s function upon the labeling. While the discovery and implementation of fluorescent proteins revolutionized live cell imaging, their poor photophysical performance (fast photobleaching/low brightness) and rather large size are still limiting factors for many applications. In contrast, a broad range of small organic fluorescent dyes with superior photophysical properties offers a better alternative for labeling. Traditional bio-coupling with fluorescent dyes employs accessible naturally occurring reactive groups (thiols or amines) of proteins and corresponding reactive fluorophore derivatives (maleimid or NHS-ester, Fig. 1a).
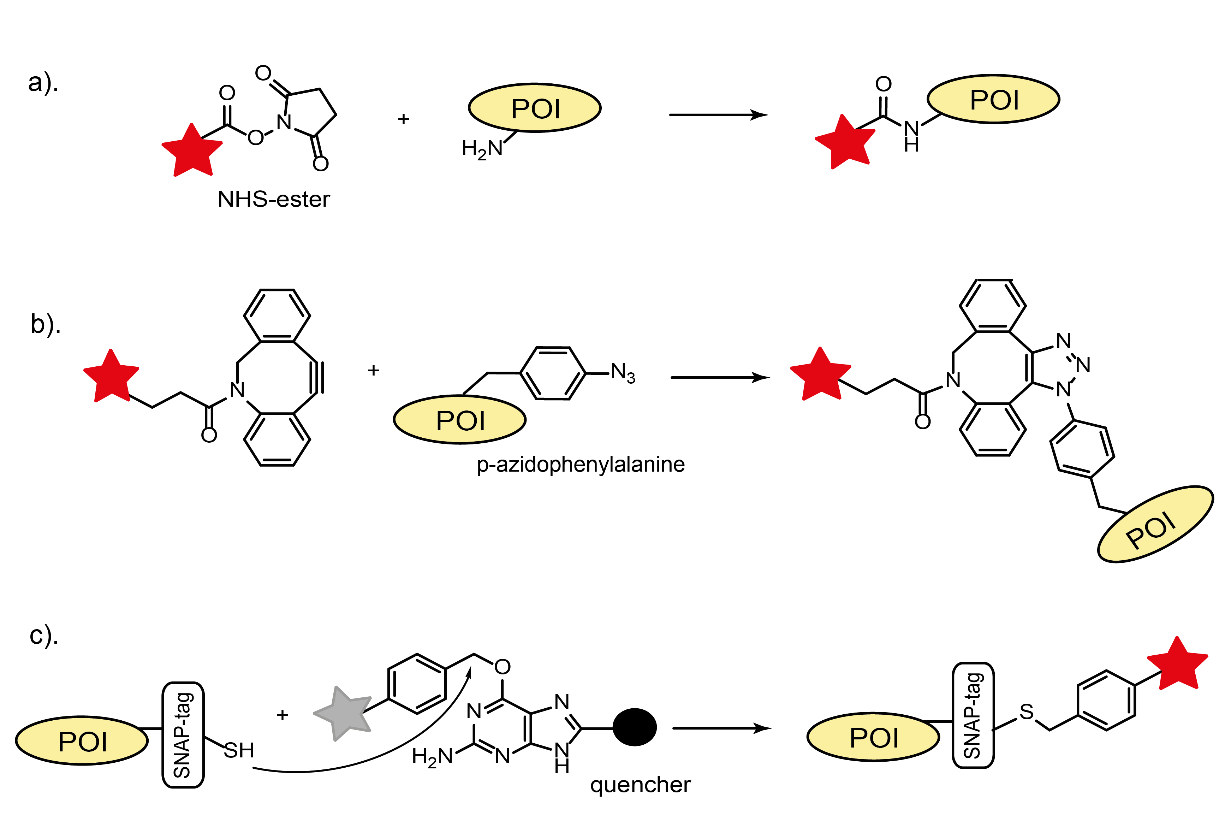
This direct conjugation of POIs with dyes is often related to in vitro investigations and to labeling of antibodies for immunostaining. A recently developed bio-orthogonal coupling strategy enables a better control over the site-specificity and labeling stoichiometry. It is based on the incorporation of unnatural amino acids, carrying unique chemical groups (e.g. para-acetyl or para-azide moieties), during the protein synthesis. These groups react very specifically with their counterparts in oxime condensations or click chemistry reactions. Additionally, a strain-promoted copper-free click reaction facilitates in vivo applications (Fig. 1b). The SLP technology (Halo-, SNAP-, and CLIP-tags) combines the advantages of all three labeling methods described above. It is based on the formation of a specific covalent bond between a reactive fluorophore derivative and a SLP-tag of fused POIs (Fig. 1c). This technique is applicable for both live and fixed cells. Recent development of so-called fluorogenic labels expanded even further the fluorescent tool-kit. A hallmark of these labels is their ability to generate fluorescence upon binding to a specific target in vivo and in vitro. This leads to a higher imaging contrast due to the reduced unspecific background. Fluorogenicity can be realized by quenching effects (PET or FRET) of the neighboring moieties that are cleaved or undergo a structural change upon labeling reaction (Fig. 1c). The coupling chemistry of unnatural amino acids or self-labeling tags can be utilized for the fluorogenic protein labeling. Depending on the application, a combination of all these labeling methods can be a method of choice.
contact
GATTAquant GmbH
Staffelseestraße 8
DE-81477 München
Phone: +49 89 2153 720 80
INFO@GATTAQUANT.COM
funded by

Payment Options


 English
English German
German Chinese
Chinese